Project 1: Patterned Heterogeneity in Colon Cancer
PIs: Christopher C.W. Hughes, Marian Waterman; Key Personnel: Steven D. Allison, Michelle Digman, Robert A. Edwards, Kai Kessenbrock, Arthur Lander, John Lowengrub, Qing Nie
Solid tumors are complex masses of cancer cells with a multitude of genetic, epigenetic, morphologic and metabolic phenotypes. This heterogeneous condition is a formidable barrier to treating cancer as it underlies the ability of tumors to adapt to nutrient starvation, immune challenges and to develop resistance to cancer treatments – the most common cause of mortality. Non-genetic heterogeneity in gene expression, signaling and metabolism are considered to be some of the most dynamic forms of heterogeneity and the most responsive to the tumor microenvironment. But we currently understand little about such heterogeneity, both mechanistically (what drives it) and functionally (how it helps the tumor). Non-genetic heterogeneity is very challenging to study, partly because of a limited toolbox and partly for lack of tractable model systems. Consequently, there are fundamental unknowns about how such heterogeneity arises and what its role in tumor growth and drug resistance really is.
In preliminary and published work, we observed heterogeneity in a xenograft model of colon cancer where the heterogeneity is patterned in a manner suggestive of a spatially self-organizing process (such as Turing-patterning). What is heterogeneous in these tumors is both Wnt signaling (thought to be the essential driver of proliferation in these cells), and metabolism (the balance between glycolysis and oxidative phosphorylation). In particular, the pattern consists of cell clusters, or spots, in which biomarkers of Wnt signaling are higher than in surrounding regions. These spots also mark regions of glycolytic metabolism. These patterns are likely connected through Wnt regulation of the expression of genes that control metabolism, as identified in our earlier work, and possibly through Wnt-Turing patterning of cancer cell subpopulations. Interestingly, glycolytic heterogeneity has recently been proposed to serve as the basis for resistance to anti-angiogenic therapy, one of the most important clinical problems in colorectal cancer. The short time scale of the xenografting (14-21 days), the reproducibility of the heterogeneity across genetically identical cell lines, and sites of injection, all suggest that this heterogeneity is not genetic.
In this project, Hughes, Waterman and their team seek to understand the causes of the observed heterogeneity in colon cancer, the reason why spatial patterns of heterogeneity develop spontaneously, the consequences of such heterogeneity for the growth of tumor cells, and whether this, or possibly other, forms of heterogeneity indeed drive resistance to therapy (and if so, why). They are addressing these questions by 1) explaining the relationships between heterogeneity, patterning and growth of colon tumors; 2) defining the general principles linking heterogeneity, patterning and growth in
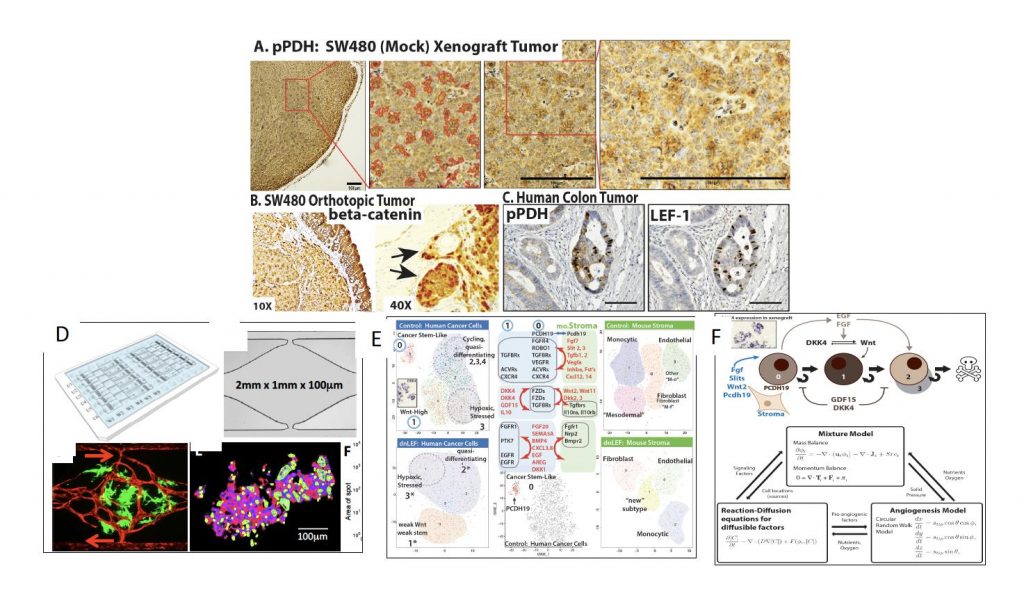
Figure 1. Xenograft colon tumors reveal a spotted pattern of metabolic (A) and Wnt signaling (B) heterogeneity; concordant heterogeneity in metabolism and Wnt signaling is present in primary patient tumors (C). (D) shows a new vascularized microtumor (VMT) platform that supports human vessels (red) and colon tumors (green), NADH-fluorescence lifetime imaging (FLIM) of colon tumors in the VMT shows metabolic heterogeneity (D: lower right). (E) Single Cell Sequence analysis using Seurat/tSNE clustering reveals that SW480 xenograft tumors are strikingly heterogeneous with a cancer stem cell-like cluster (0: top left) that is missing in tumors expressing dominant negative LEF1 (dnLEF: bottom left). In (E/Middle Panel) Ligand and receptor pairs among several cell clusters of the human and mouse (right panels in E) cells are shown. (F) Candidate mathematical model for simulating SW480 tumors.
colon tumors; and 3) defining the link between heterogeneity and drug resistance.
The foundation of the project is multi-scale modeling of stochastic and self-organizing processes that potentially explains overt differences in tumor growth, patterning of heterogeneity and metabolism, and the most likely mechanisms for drug resistance. The experimental tools pair xenograft studies with a novel platform for generating fully vascularized micro-tumors in vitro; the use of fluorescence lifetime imaging to read out the metabolic states of unlabeled, living cells, and the use of single cell transcriptomics
to identify cell states, the gene expression signatures that define them and signaling and adhesion molecules that mediate communication among the cells. Modeling predictions of strategies that re-establish drug sensitivity will be tested via genetic engineering (CRISPR/Cas9) or small molecule drug therapies. The overarching goal of the work is to discover deep insights into the origins and consequences of tumor heterogeneity in an especially manipulable, and clinically relevant tumor system. The integration of this work with Projects 2 and 3, which focus on different cancer types (melanoma and chronic myeloid leukemia, respectively), enables us to identify general principles that underlie how the in vivo behaviors of transformed cells are constrained by rules inherited from the communities of diverse, interacting cell types and lineage hierarchies within which those cells arise.