The overall goal for the UCI Center for Cancer Systems Biology (CaSB@UCI), also known as the Center for Complexity, Cooperation and Community in Cancer, is to understand the principles that underlie why cancers are organized as they are. Our approaches stem from the idea that cancer cells proliferate and evolve in complex environments that have been highly selected for the robust control of growth and differentiation, and thus the behaviors of cancer cells can only be fully understood in the context of the design principles underlying such control. CaSB@UCI is a joint effort by the systems biology and cancer biology communities at the University of California Irvine (UCI), as represented by two campus-wide research organizations, the Center for Complex Biological Systems and the Cancer Research Institute of the UCI Chao Family Comprehensive Cancer Center.
Funding for the center comes from the National Cancer Institute, through the Cancer Systems Biology Centers program.
Research Overview
The UCI Center for Cancer Systems Biology (CaSB@UCI) is carrying out three coordinated, team-oriented research projects on the role of context, cooperation and community in the initiation and progression of cancer. All three projects seek to understand how the in vivo behaviors of transformed cells are constrained by rules inherited from the communities of diverse, interacting cell types and lineage hierarchies within which those cells arise...
PIs: Christopher C.W. Hughes, Marian Waterman; Key Personnel: Steven D. Allison, Michelle Digman, Robert A. Edwards, Kai Kessenbrock, Arthur Lander, John Lowengrub, Qing NieSolid tumors are complex masses of cancer cells with a multitude of genetic, epigenetic, morphologic and metabolic phenotypes. This heterogeneous condition is a formidable barrier to treating cancer as it underlies the ability of tumors to adapt to nutrient starvation, immune challenges and to develop resistance to cancer treatments – the most common cause of mortality. Non-genetic heterogeneity in gene expression, signaling and metabolism are considered to be some of the most dynamic forms of heterogeneity and the most responsive to the tumor microenvironment. But we currently understand little about such heterogeneity, both mechanistically (what drives it) and functionally (how it helps the tumor). Non-genetic heterogeneity is very challenging to study, partly because of a limited toolbox and partly for lack of tractable model systems. Consequently, there are fundamental unknowns about how such heterogeneity arises and what its role in tumor growth and drug resistance really is.In preliminary and published work, we observed heterogeneity in a xenograft model of colon cancer where the heterogeneity is patterned in a manner suggestive of a spatially self-organizing process (such as Turing-patterning). What is heterogeneous in these tumors is both Wnt signaling (thought to be the essential driver of proliferation in these cells), and metabolism (the balance between glycolysis and oxidative phosphorylation). In particular, the pattern consists of cell clusters, or spots, in which biomarkers of Wnt signaling are higher than in surrounding regions. These spots also mark regions of glycolytic metabolism. These patterns are likely connected through Wnt regulation of the expression of genes that control metabolism, as identified in our earlier work, and possibly through Wnt-Turing patterning of cancer cell subpopulations. Interestingly, glycolytic heterogeneity has recently been proposed to serve as the basis for resistance to anti-angiogenic therapy, one of the most important clinical problems in colorectal cancer. The short time scale of the xenografting (14-21 days), the reproducibility of the heterogeneity across genetically identical cell lines, and sites of injection, all suggest that this heterogeneity is not genetic.In this project, Hughes, Waterman and their team seek to understand the causes of the observed heterogeneity in colon cancer, the reason why spatial patterns of heterogeneity develop spontaneously, the consequences of such heterogeneity for the growth of tumor cells, and whether this, or possibly other, forms of heterogeneity indeed drive resistance to therapy (and if so, why). They are addressing these questions by 1) explaining the relationships between heterogeneity, patterning and growth of colon tumors; 2) defining the general principles linking heterogeneity, patterning and growth in
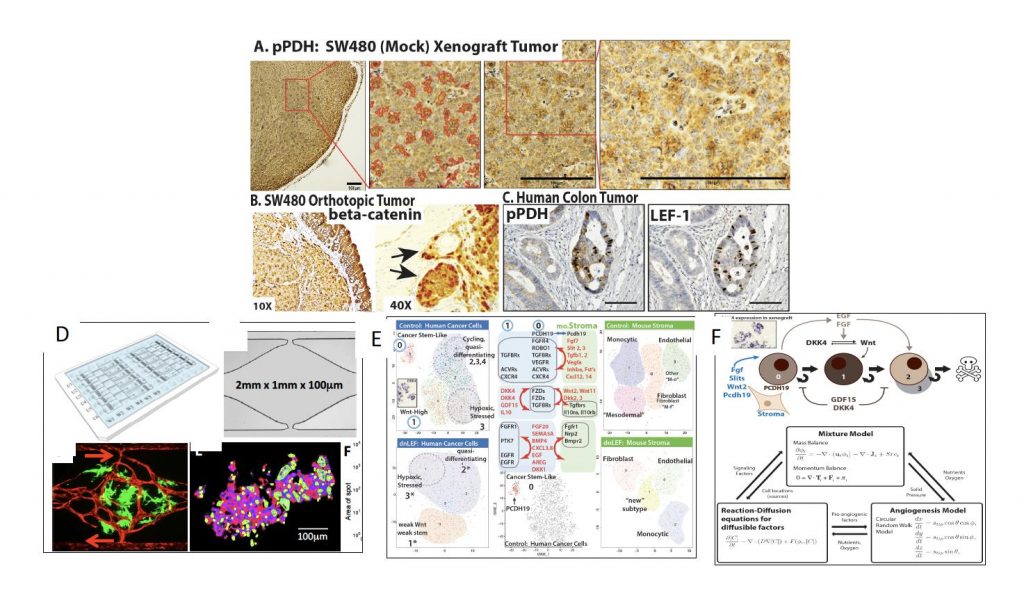 Figure 1. Xenograft colon tumors reveal a spotted pattern of metabolic (A) and Wnt signaling (B) heterogeneity; concordant heterogeneity in metabolism and Wnt signaling is present in primary patient tumors (C). (D) shows a new vascularized microtumor (VMT) platform that supports human vessels (red) and colon tumors (green), NADH-fluorescence lifetime imaging (FLIM) of colon tumors in the VMT shows metabolic heterogeneity (D: lower right). (E) Single Cell Sequence analysis using Seurat/tSNE clustering reveals that SW480 xenograft tumors are strikingly heterogeneous with a cancer stem cell-like cluster (0: top left) that is missing in tumors expressing dominant negative LEF1 (dnLEF: bottom left). In (E/Middle Panel) Ligand and receptor pairs among several cell clusters of the human and mouse (right panels in E) cells are shown. (F) Candidate mathematical model for simulating SW480 tumors.colon tumors; and 3) defining the link between heterogeneity and drug resistance.The foundation of the project is multi-scale modeling of stochastic and self-organizing processes that potentially explains overt differences in tumor growth, patterning of heterogeneity and metabolism, and the most likely mechanisms for drug resistance. The experimental tools pair xenograft studies with a novel platform for generating fully vascularized micro-tumors in vitro; the use of fluorescence lifetime imaging to read out the metabolic states of unlabeled, living cells, and the use of single cell transcriptomics
Figure 1. Xenograft colon tumors reveal a spotted pattern of metabolic (A) and Wnt signaling (B) heterogeneity; concordant heterogeneity in metabolism and Wnt signaling is present in primary patient tumors (C). (D) shows a new vascularized microtumor (VMT) platform that supports human vessels (red) and colon tumors (green), NADH-fluorescence lifetime imaging (FLIM) of colon tumors in the VMT shows metabolic heterogeneity (D: lower right). (E) Single Cell Sequence analysis using Seurat/tSNE clustering reveals that SW480 xenograft tumors are strikingly heterogeneous with a cancer stem cell-like cluster (0: top left) that is missing in tumors expressing dominant negative LEF1 (dnLEF: bottom left). In (E/Middle Panel) Ligand and receptor pairs among several cell clusters of the human and mouse (right panels in E) cells are shown. (F) Candidate mathematical model for simulating SW480 tumors.colon tumors; and 3) defining the link between heterogeneity and drug resistance.The foundation of the project is multi-scale modeling of stochastic and self-organizing processes that potentially explains overt differences in tumor growth, patterning of heterogeneity and metabolism, and the most likely mechanisms for drug resistance. The experimental tools pair xenograft studies with a novel platform for generating fully vascularized micro-tumors in vitro; the use of fluorescence lifetime imaging to read out the metabolic states of unlabeled, living cells, and the use of single cell transcriptomicsto identify cell states, the gene expression signatures that define them and signaling and adhesion molecules that mediate communication among the cells. Modeling predictions of strategies that re-establish drug sensitivity will be tested via genetic engineering (CRISPR/Cas9) or small molecule drug therapies. The overarching goal of the work is to discover deep insights into the origins and consequences of tumor heterogeneity in an especially manipulable, and clinically relevant tumor system. The integration of this work with Projects 2 and 3, which focus on different cancer types (melanoma and chronic myeloid leukemia, respectively), enables us to identify general principles that underlie how the in vivo behaviors of transformed cells are constrained by rules inherited from the communities of diverse, interacting cell types and lineage hierarchies within which those cells arise.
Project 2: Understanding the Cellular Origins of Melanoma
PIs: Anand Ganesan, Arthur Lander; Key Personnel: Devon A. Lawson, John Lowengrub, Bruce Tromberg, Tatiana Krasieva
Melanoma, a tumor resistant to therapy in late stages, is curable by excision when caught early. Early melanomas can be difficult to distinguish from benign, pigmented “moles”, i.e. melanocytic nevi; this leads to unnecessary excision of many normal nevi while early melanomas are often missed. Nevi and melanomas share more than morphological features: Clinical and experimental data show that ~90% of nevi are initiated when melanocytes acquire an activating mutation in the BRAF oncogene, the same oncogenic mutation observed in >60% of melanomas. Yet nevi spontaneously stop growing. This is usually attributed to “oncogene-induced senescence,” but the fact that nevi readily re-grow after incomplete excision, or in response to UV-irradiation, and can sometimes evolve to melanoma, suggest nevi are not “senescent” but reversibly growth-arrested. Nevi also spontaneously regress, a process that appears to involve the immune system.
In preliminary work, we investigated nevus dynamics in a mouse model of inducible Braf activation, which mimics human nevus formation and also produces melanomas either at low frequency or when additional oncogenic mutations are added (e.g., in Pten). We found that as we activate Braf in more melanocytes, such that nevi become more numerous and closely-spaced, the smaller individual nevi become—as if nevi, when close together enough, inhibit each other’s growth. Such behavior is predicted by mathematical models of growth control based on feedback through diffusible signaling molecules. Such models achieve robust control when feedback regulates decisions between self-renewal and progression to alternate cell states or fates. Interestingly, when we look closely at the nevi in this model, we see that there are, in fact, two distinct cell types: highly pigmented nevus body cells and a scattered, lightly pigmented melanocyte population that forms a “veil” around the pigmented cells that had not been observed before. These veil cells are usually not seen unless the melanocyte lineage is fluorescently-labeled with GFP. Single cell RNA-sequencing suggests that these cells likely communicate through ligands and receptors they differentially express.
In this project, Ganesan, Lander and their team seek to understand the role of the nevus body and veil cell types in mouse models that produce both nevi and melanoma,
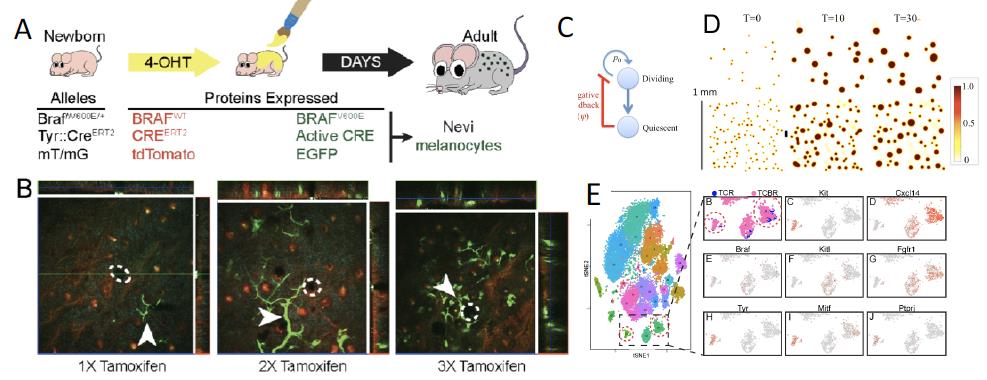
Figure 2. (A) BRAF-mutant (Tyrosinase::CreERT2; BraffloxV600E/+) mice are crossed with ROSAmT/mG mice (TCBR) to GFP-label BRAF-mutant melanocytes. (B) Fluorescence emission (confocal and MPM) 3-D imaging of skin of live mice (as in Fig. 1; total depth = 90, 115 and 190μm for 1x, 2x, and 3x representative stacks, respectively). Green = GFP; red = td Tomato. (C,D). Continuum modeling, in which cells at an arrested (quiescent) lineage stage feedback on the self-renewal probability of dividing cells. Model snapshots (D) show cell density (see heatmap) as a function of time (T= cell cycles) and location. Top and bottom rows are for low and high seeding densities, respectively, that represent different levels of nevi induction. (E) Single Cell RNA-seq of melanocytic nevi. Control mice (Tyrosinase::CreERT2; ROSAmTmG; TCR) and TCBR mice, as labeled. Left: tSNE clustering of gene expression for individual cells. Right: A portion of the tSNE map, showing contributions of TCR and TCBR mice as well as expression of selected marker genes.
and to identify both the nature of how growth is controlled in nevi and the means by which melanoma cells escape from it. The goal of the work is to build a solid molecular and cellular framework on which to base clinical decisions about melanoma prevention, detection and treatment. They are addressing these issues by 1) Explaining feedback growth dynamics in melanocytic nevi; 2) Elucidating how melanomas escape growth control mechanisms that arrest nevi; and 3) Revealing how the immune system targets nevi, and how this affects melanoma development.
This project integrates multiscale mathematical modeling with experiments in mice using a nevus-forming inducible activated Braf model, and a version of the same model that combines Braf activation with inducible loss of one allele of Pten, leading to the reliable production of both nevi and melanoma tumors. We are developing hypotheses that can explain the spatiotemporal dynamics and spatial statistics of nevus and melanoma development in these models, including potential bifurcations that account for the development of both nevi and melanoma in the same mouse. We are investigating the reasons why some cells escape from growth control, while others do not. We anticipate that this is unlikely to be due to a requirement for inactivation of the other Pten allele, and instead believe that escape may more likely be due to the spatial dynamics of collective feedback. The results are expected to shed light on signaling pathways that could be manipulated to prevent or treat melanoma. Live cell imaging, focused laser ablation, immunohistochemistry, and time-course single cell RNA-sequencing are used to identify potential positive and negative feedback regulators that drive the mathematical models, and experiments are used to test model-based predictions concerning the roles that such molecules play. Finally, an investigation of spontaneous regression, which occurs with both mouse and human nevi, provides clues into how the immune system efficiently recognizes melanocyte overgrowth. Since immunotherapy has recently emerged as a promising therapy for melanoma, this study is expected to reveal whether immunotherapy leverages an existing immune program for eliminating nevi, and if so, how that program is carried out, and how melanomas typically escape from it. Such information should aid in developing new prevention and therapeutic strategies for this devastating disease. The integration of this work with Projects 1 and 3 occurs through the use of scRNA-seq as a tool for hypothesis generation and development and through the application of mathematical models that have similar underlying structures (cell state transitions, proliferation and quiescence, positive and negative feedback), but differ in their context (e.g., spatial in Projects 1 and 2 vs. non-spatial in Project 3).
Project 3. Modeling malignant myelopoiesis to increase efficacy of targeted leukemia therapy
PI: Richard Van Etten; Key Personnel: Kai Kessenbrock, Natalia Komarova, John S. Lowengrub, Qing Nie, Dominik Wodarz, Xiaohui Xie, Nilamani Jena
Chronic myeloid leukemia (CML), one of the most prevalent of human leukemias, is a natural model of dysregulated granulopoiesis driven by a single genetic abnormality in a hematopoietic stem cell, the BCR-ABL1 gene fusion. Although therapy with tyrosine kinase inhibitors (TKIs) such as imatinib mesylate has dramatically lowered the death rate in CML, lifelong treatment is needed and the associated economic costs are significant. Two major unaddressed questions are to understand the mechanism of primary resistance to TKI therapy (affecting ~10-15% of newly diagnosed CML patients), and to identify strategies to increase the frequency of complete molecular remission (CMR) in patients treated with TKIs and subsequently the rate of treatment-free remission (TFR), which may represent a surrogate for permanent cure of the disease. The scientific premise of this project is that new and clinically relevant insights into the biology of CML and its response to therapy can be gained by a more physiologically accurate mathematical model of the disease.
While mathematical models of CML have been developed by several groups, these models tend to be highly simplified and largely omit feedback interactions among the different hematopoietic components. These models are designed to fit clinical data sets of the response of patient populations to TKI therapy but unfortunately, they have not proven useful for understanding primary TKI resistance or for predicting TFR. In preliminary work, we found that nonlinear models that incorporate feedback are more robust, have a better fit to alternative patterns of patient response than simple linear models used previously, and allow predictions about the effects of interventions affecting parameters (such as cell cycle status) subject to feedback mechanisms on the response to TKI therapy. Preliminary analyses from prototype feedback models have already raised two provocative hypotheses about CML. The first is that the initial response to TKI therapy may depend on the relative size of the leukemic stem cell clone, which will be analyzed by TKI treatment of mice engrafted with different levels of BCR-ABL1+ stem cells. The second is that interventions that increase leukemic stem cell cycling may sensitize this population to killing by TKIs.
In this project, Van Etten and his team seek to develop improved mathematical models of chronic phase CML and the response to TKI therapy, to validate these models using data from a binary transgenic mouse model of CML and from human CML patients, and to utilize the models to test several hypotheses about the response of CML to therapy and to predict strategies for improving the treatment-free remission rate in CML. We address these issues by 1) Developing data-driven, dynamic models of CML hematopoiesis incorporating feedback control; 2) Measuring granulocytopoiesis parameters in a CML mouse model and in human CML patients; 3) Testing model-driven hypotheses about the response of CML to therapy and informing strategies for improving the treatment-free remission rate in CML.
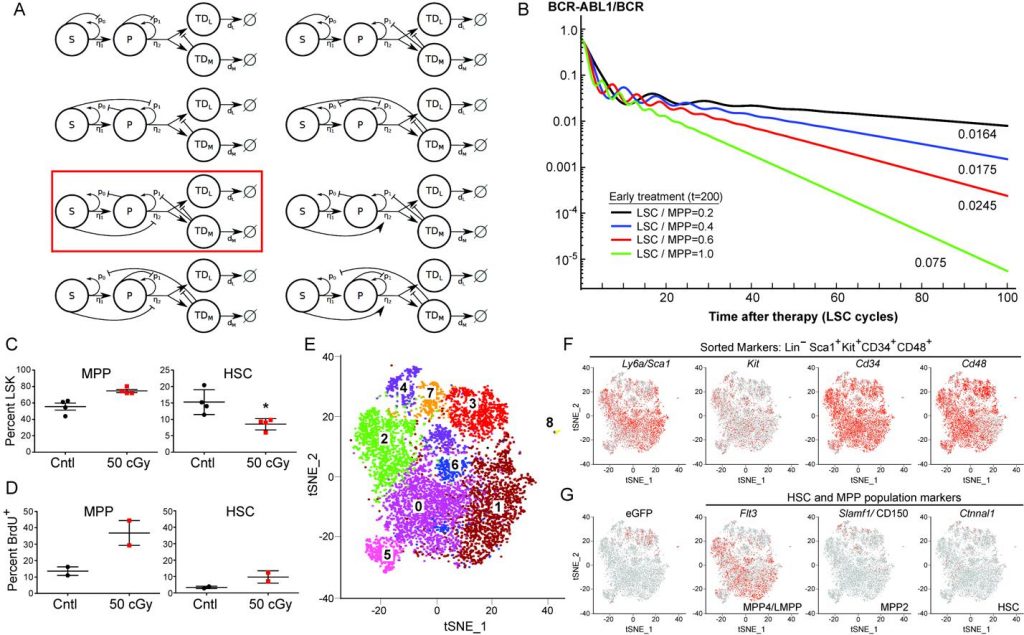
Figure 3. (A) Illustrative nonlinear feedback models of hematopoiesis. The red box indicates the model used in (B). (B) Increased leukemic stem cell (LSC) cycling predicts faster LSC decline on TKI therapy. The results correspond to different LSC cycling parameters applied (expressed as ratio of LSC to multipotential progenitor (MPP) cycling rate). Note the increasing negative slope of the second linear phase with increased cycling. (C,D) Selective decrement of HSC compartment increases MPP proliferation. Mice were irradiated (50 cGy), injected 24h post-radiation with BrdU and analyzed 12h later. (C) 50 cGy irradiation significantly (P=0.018) decreased the size (%LSK) of the HSC compartment (right panel) at 36h without a significant effect on the MPP compartment (left panel). (D) Reduction in HSC pool by low-dose radiation dramatically increases proliferation of the MPP compartment, supporting the existence of a negative feedback loop in the model above. (E) t-SNE analysis of scRNA-seq data from the MPP population isolated from mice with BCR-ABL1-induced CML shows eight separate subpopulations. (F) Expression of sorted lineage markers mapped on the t-SNE plot from (E). (G) Expression of GFP (marking leukemic MPP) and MPP/HSC markers mapped onto the t-SNE plot from (E).
Machine-based automated model selection methods are being utilized to arrive at a mathematical model that maintains appropriate stability and homeostasis, responds physiologically to stress and depletion of different cell compartments, and conforms to the limited existing qualitative data on CML hematopoiesis derived from mouse models and patient studies. To validate and inform potential models, binary/conditional BCR-ABL1 transgenic donor mice are being used to generate mixed BM chimeras via transplantation of high doses of unfractionated marrow cells without use of conditioning radiation. Recipients bearing a clone of BCR-ABL1+ cells have leukemia induced by withdrawal of doxycycline, leukemic mice are treated with BrdU or subjected to short-term stable isotope labeling with D2-glucose, marrow and spleen stem and progenitor compartments isolated by flow cytometry, and cell cycle and kinetic parameters estimated to inform the mathematical models. Single cell transcriptome profiling is used to investigate the heterogeneity of the MPP compartment and to discover potential regulatory mechanisms mediated by cytokine/receptor interactions. Feedback relationships are tested via direct in vivo manipulation using depleting monoclonal antibodies and through a novel mouse that targets metronidazole cytotoxicity to specific cell compartments. Parallel studies of human CML progenitor flux will be carried out through a clinical protocol of short-term D2-glucose labeling in patients presenting with suspected CML prior to diagnostic BM biopsy. The response to TKI therapy is also being analyzed. The integration of this work with Projects 1 and 2 occurs through the use of scRNA-seq as a tool for hypothesis generation and development and through the application of nonlinear mathematical models that have similar underlying structures (cell state transitions, proliferation and quiescence, positive and negative feedback), although the models in Project 3 are non-spatial.
All three projects combine mathematical modeling, genomics, and experimental manipulation of animal models. Mathematical modeling is central to all three projects, not just as a means to analyze large data sets, but as a way of identifying, with the most generality, the architectures of cell interaction and feedback that can explain generic features of cancer cell behavior. All three projects develop models with design principles built on similar concepts: cell state transitions, proliferation and quiescence, positive and negative feedback, but in different contexts that derive from the different cancer types (spatial vs. non-spatial; focus on self-organizing pattern vs. focus on control; emphasis on model and parameter identification vs. emphasis on replicating qualitative behaviors). A common theme is the idea that bi- and multistability that arises as a result of feedback can potentially explain bifurcating system behaviors, such as nevi and melanoma in the same mouse, spatial patterns of heterogeneity, or resistance to cancer therapy, without having to attribute such events to new mutational “hits.”
The three projects are served by the following core facilities:
Single Cell Analysis Core
Core Director: Suzanne Sandmeyer; Key personnel: Kai Kessenbrock, Devon A. Lawson, Melanie Oakes, Ali Mortazavi, Jie (Jenny) Wu
This core provides technical, bioinformatic, and training support for the Center. The goal of the Single Cell Analysis Core technical support is to make cost-effective high-throughput analysis of single cells optimal for and accessible to each of the three Projects. Technical support consists of providing staff and instrument infrastructure, as well as advising on design of experimental strategies, facilitating sharing of protocols for process development; providing single-cell services including microscopy, protein localization, cell sorting, library production, and sequencing; and working with investigators to innovate in these technologies.